Symbiotic nitrogen-fixing rhizobia enable plant growth in poor soils
Due to their ability to fix nitrogen in symbiosis with legumes, rhizobia play an important role in agriculture. They enable growth of legumes on low-nitrogen soils without additional nitrogen supply. The root nodule forming bacterium Sinorhizobium meliloti living in symbiosis with alfalfa was chosen for analysis. Potent genetic methods and techniques for engineering were developed for S. meliloti allowing a detailed analysis of this species. In the area of genome research, S. meliloti was one of the first bacteria to be completely sequenced. The genome sequence of S. meliloti 1021 was established within an international consortium including European, Canadian and US-American researchers (Galibert et al., Science 293, 2001: 668 - 672). Subsequently, the genome sequences of further Sinorhizobium strains were analyzed at the CeBiTec, namely Sinorhizobium fredii HH103, Sinorhizobium meliloti RM41 and Sinorhizobium meliloti SM11. These sequencing projects were supervised by S. Weidner .
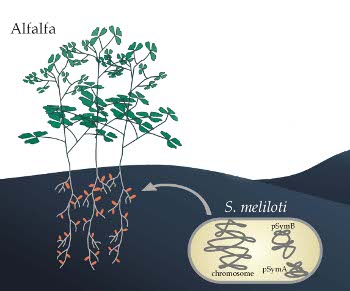
Fig. 1: Symbiotic nitrogen fixation between the legume lucerne and the soil bacterium Sinorhizobium meliloti
The soil bacterium Sinorhizobium meliloti 1021 induces root nodules on alfalfa. After colonizing these root nodules, S. meliloti cells differentiate into nitrogen-fixing bacteroids. The genome of S. meliloti 1021 comprises a chromosome and the two megaplasmids pSymA and pSymB.
Amino acids produced by corynebacteria are used as feed additives for animal nutrition
Corynebacterium glutamicum produces the amino acid lysine which is used as a feed additive for animal nutrition worldwide. In cooperation with the company Degussa, now Evonik Industries AG, the strain Corynebacterium glutamicum ATCC 13032 was genetically analyzed and finally sequenced (Kalinowski et al., J. Biotechnol. 104, 2003: 5 – 25). After establishing the genome sequence, amino acid production was improved by using omics technologies. C. glutamicum is very well suited for genome-based systems biology and is regarded as a platform organism for Synthetic Biology. The analyses mentioned above were carried out in close cooperation with J. Kalinowski and his group.
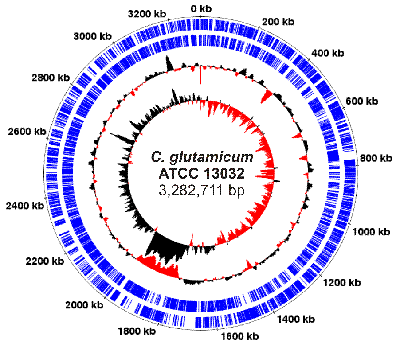
Fig. 2: Genome plot of Corynebacterium glutamicum ATCC 13032
The genome of C. glutamicum ATCC 13032 comprises a circular chromosome with a size of 3.2 million bases and about 3,000 protein-coding genes. The genome was sequenced from 1999-2001. In 2003, it was published by J. Kalinowski et al. in Journal of Biotechnology.
Xanthan-synthesizing xanthomonads produce a versatile thickening agent
Xanthomonas campestris pv. campestris (Xcc), a GRAS organism (GRAS: generelly regarded as safe), is used for the industrial production of the exopolysaccharide xanthan. Xanthan serves as thickener for food, but can also be found in pharmaceuticals and cosmetic products. The genome of the xanthan-producing Xcc B100 was sequenced (Vorhölter et al., Journal of Biotechnol. 134, 2008: 33 – 45) and analzed by means of systems biology. At the moment, a cooperation between the company Jungbunzlauer, Vienna, Austria, and the CeBiTec of Bielefeld University aims to improve xanthan biosynthesis using omics analyses. This project is carried out in cooperation with K. Niehaus. and coordinated by the scientists V. Ortseifen and M. Ruwe.
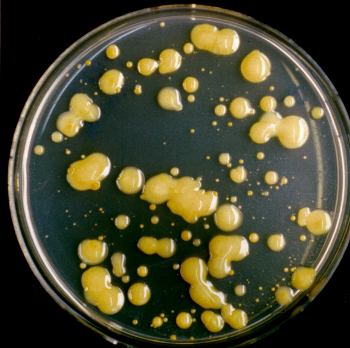
Fig. 3: Colonies of Xanthomonas campestris pv. campestris (Xcc) B100 on solid agar.
The Xcc wild type colonies are characterized by a smooth, slimy phenotype indicating the production of the exopolysaccharide xanthan.
Acarbose synthesized by Actinoplanes strains is used as therapeutic for the treatment of diabetes type II
Acarbose, a secondary metabolite produced by Actinoplanes, is used in medicine for the treatment of diabetes II. Acarbose is marketed as “Glucobay” by Bayer Pharma AG. In cooperation with the company, the genome of the acarbose-synthesizing strain Actinoplanes sp. SE50/110 was sequenced (Schwientek et al. BMC Genomics13, 2012:112). The sequence data now enables the detailed analysis by omics technologies. The project was carried out together with the group of J. Kalinowski and is coordinated by the two scientists S. Schneiker-Bekel and M. Persicke.
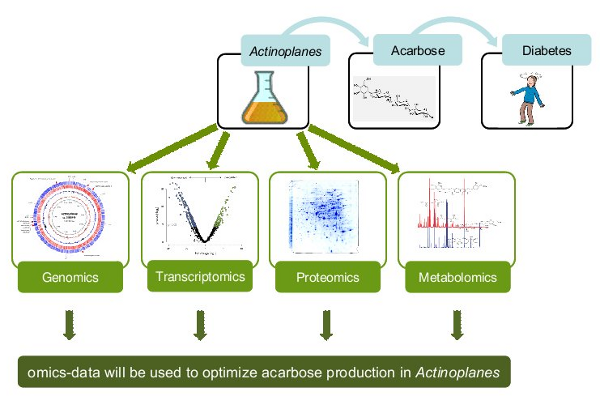
Fig. 4: Overview on omics analyses of the acarbose producer Actinoplanes sp. SE50/110
In the meantime, the four omics technologies presented were successfully established for Actinoplanes sp. SE 50/110. The genome was sequenced and the genome-wide gene expression was analyzed. Furthermore, protein localization and metabolite identification were carried out.



