EMMA
EMMA Features
As a high throughput technique, microarray experiments produce large datasets that consist of measured data, laboratory protocols, and experimental settings. Apart from efficient storage, state-of-the-art algorithms, and visualisations, the interpretation of data from microarrays often requires mapping large data-sets onto a variety of genomic data sources.
Basic Features
• MAGE-ML compatibility
• Interface with high usability
• "One-click-solutions"
• Efficient normalization
• Easy experimental setup and process control
• Applicable in distributed projects
Advanced Features
• Extensible analysis tools and plug-in architecture
• Advanced data analysis and data mining
• Integration of external Omics data sources
• Finegrained access control
• User configurable data analysis pipelines
Implementation
EMMA 2 is mainly written in Perl while portions are in R and Java. The data repository is implemented using O2DBI, which is an object-oriented code generator. O2DBI significantly simplifies the creation of complex database applications. Additionally, EMMA 2 provides role-based access control for each data-object. The application programmer's interface(API) is implemented and documented by the automatically generated Perl modules (created by O2DBI) that also contain additional manually added functions.
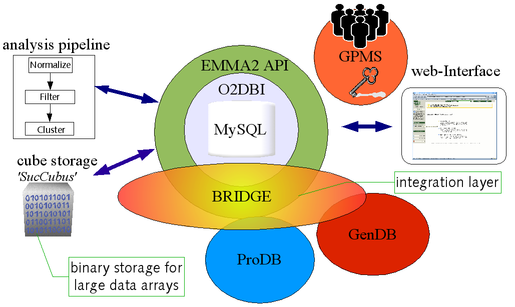
The EMMA classes are mapped onto tables via O2DBI and stored in an SQL database. All access to these data is performed via a Perl client and server API. On the client side user friendly interfaces have been implemented that simply use the functionality of these APIs.
EMMA2 Documentation
Information about both systems including developer documentation, administrator documentation and FAQs can be found in our software Wikis:
EMMA2 Access
EMMA2 demo project
You can access the EMMA2demo project via guest login here.
This project contains several demo experiments of which two are complete finished, giving a impression how experiments are represented, which typical analysis can be performed and and what results are generated. Note that as it is a demo project, you won't be able to import data from ArrayLIMS or to start to compute data using the "start analysis" option.
Invididual EMMA2 and ArrayLIMS project
If you want to work with your own data set on the CeBiTec EMMA2 and ArrayLIMS server, feel free to contact the
As soon as you have set up a project, received a username and password, you can login into EMMA2 following this link and into ArrayLIMS following this link.
System requirements for EMMA 2 and ArrayLIMS
- Please enable JavaScript, Cookies, and Pop-ups
- Java (Java-PlugIn) >= 1.4 is required to use the Cluster Viewer
- The system has been tested with Mozilla Firefox. We recommend to use a recent version



