Fusion - PolyOmics data analysis, visualization and integration

Fusion is a web-based platform for the integrative analysis of omics data providing a collection of new and established tools and visualization methods to support researchers in exploring multi-level omics data.
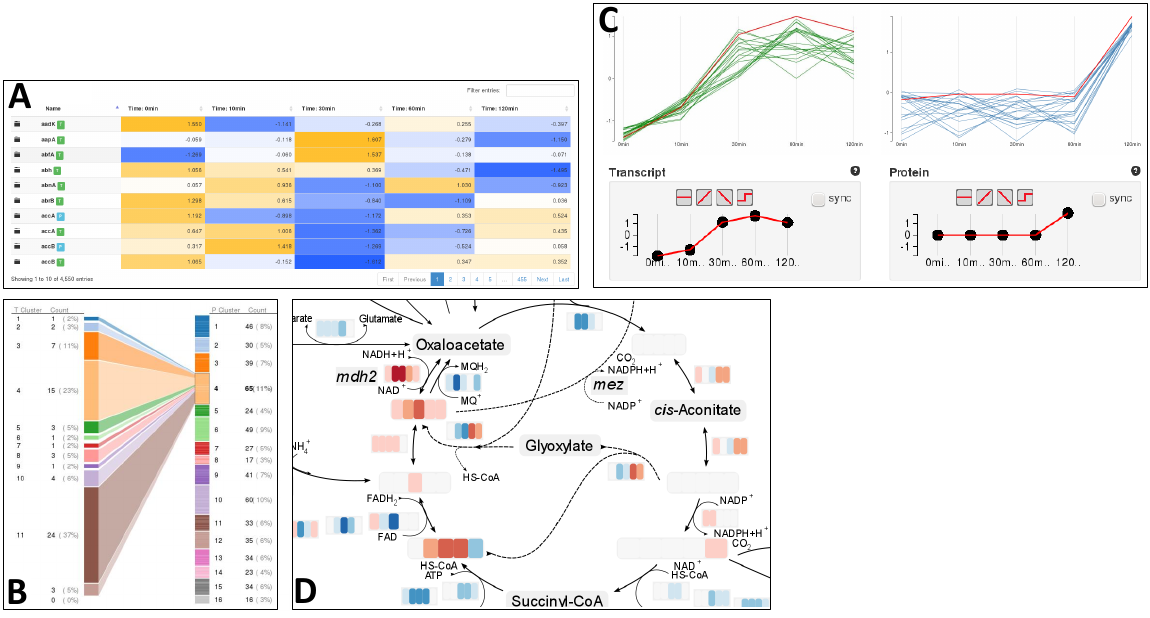
High-throughput experimental technologies transformed biological research from a relatively data-poor discipline to one that is data-rich. The challenge is no longer to generate experimental data, but to compute and analyze it. A key aspect of understanding and analyzing data is visualization. A powerful visualization can make the difference whether the user is able to gain a mental model for his data and apply his biological knowledge. Furthermore, to fully understand a biological metabolism and its responses to environmental factors, it is necessary to include functional characterization and accurate quantification of all levels - gene products, proteins and metabolites, as well as their interactions.
Fusion has been developed to cope with this challenge and offers powerful analysis tools, including established methods for analyzing and visualizing single omics data, as well as new features for an integrative analysis of data from multiple omics sources. Fusion is supposed to supersede ProMeTra including its core functionality.
Interested in Omics Fusion?
Usage is free of charge. Please go to the Fusion homepage and register for an account or contact us at
Brink, B., Seidel, A., Kleinbölting, N., Nattkemper, T. W., & Albaum, S. 2016. "Omics Fusion - A Platform for Integrative Analysis of Omics Data". Journal of Integrative Bioinformatics 13:296.
Neuweger, H., M. Persicke, S. Albaum, T. Bekel, M. Dondrup, A.T. Hüser, J. Winnebald, J. Schneider, J. Kalinowski, & A. Goesmann. 2009. “Visualizing post genomics data-sets on customized pathway maps by ProMeTra – aeration-dependent gene expression and metabolism of Corynebacterium glutamicum as an example”. BMC Systems Biology, 3:82.
PUB | PubMed



