In the meantime, next generation sequencing enables the metagenome analysis of microbial communities with reasonable efforts. Bioinformatics tools play a dominant role due to the fact that each read of a high-throughput sequencing run has to be assessed taxonomically and functionally. In the meantime, single reads can be assembled to contigs, and metagenomically assembled genomes (MAGs) can be constructed by binning of contigs belonging to a particular species. In addition to metagenome analyses, metatranscriptome and metaproteome analyses have been performed.
Within the Senior Research Group, metagenome research was established using the example of microbial communities residing in biogas plants. In the meantime, a variety of projects in the field of biogas production are carried out in cooperation with several groups in Germany. These projects are for example financed by the Agency for Renewable Resources (Federal Ministry of Food and Agriculture, BMEL) and the Federal Ministry of Education and Research (BMBF).
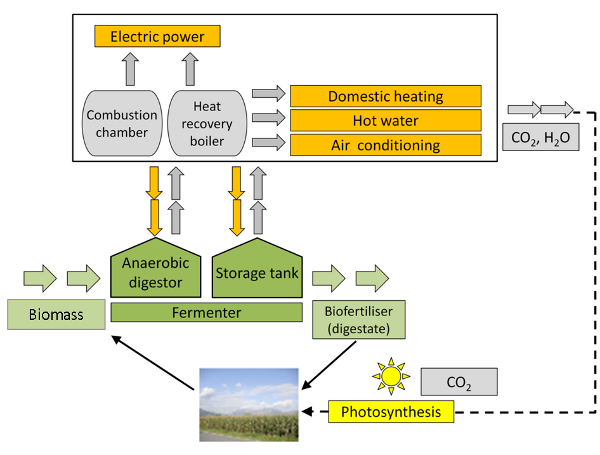
Fig. 5: From biomass to biogas to power and heat generation
Aside from metagenome analyses of microbial communities of biogas fermenters, microbial communities from silage fermentation, but also from agricultural soils were analyzed. All these metagenome experiments were carried out by Ph.D. students and supervised by A. Schlüter. A. Sczyrba and his group were involved in the bioinformatic analysis of data produced by metagenomics and metatranscriptomics experiments.



