As a high throughput technique, microarray experiments produce large datasets that consist of measured data, laboratory protocols, and experimental settings. Apart from efficient storage, state-of-the-art algorithms, and visualisations, the interpretation of data from microarrays often requires mapping large data-sets onto a variety of genomic data sources.
Basic Features
• MAGE-ML compatibility
• Interface with high usability
• "One-click-solutions"
• Efficient normalization
• Easy experimental setup and process control
• Applicable in distributed projects
Advanced Features
• Extensible analysis tools and plug-in architecture
• Advanced data analysis and data mining
• Integration of external Omics data sources
• Finegrained access control
• User configurable data analysis pipelines
Implementation
EMMA 2 is mainly written in Perl while portions are in R and Java. The data repository is implemented using O2DBI, which is an object-oriented code generator. O2DBI significantly simplifies the creation of complex database applications. Additionally, EMMA 2 provides role-based access control for each data-object. The application programmer's interface(API) is implemented and documented by the automatically generated Perl modules (created by O2DBI) that also contain additional manually added functions.
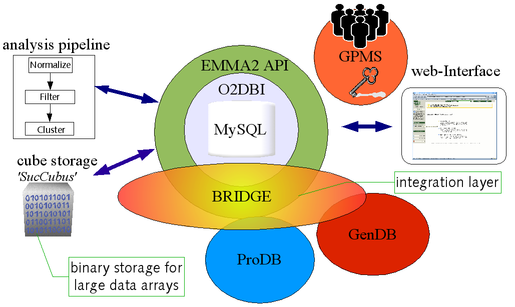
The EMMA classes are mapped onto tables via O2DBI and stored in an SQL database. All access to these data is performed via a Perl client and server API. On the client side user friendly interfaces have been implemented that simply use the functionality of these APIs.



